Agglomerative Clustering in Machine Learning
Agglomerative clustering is a hierarchical clustering algorithm that starts with each data point as its own cluster and iteratively merges the closest clusters until a stopping criterion is reached. It is a bottom-up approach that produces a dendrogram, which is a tree-like diagram that shows the hierarchical relationship between the clusters. The algorithm can be implemented using the scikit-learn library in Python.
Agglomerative Clustering Algorithm
Agglomerative Clustering is a hierarchical algorithm that creates a nested hierarchy of clusters by merging clusters in a bottom-up approach. This algorithm includes the following steps −
- Treat each data point as a single cluster
- Compute the proximity matrix using a distance metric
- Merge clusters based on a linkage criterion
- Update the distance matrix
- Repeat steps 3 and 4 until a single cluster remains
Why use Agglomerative Clustering?
The Agglomerative clustering allows easy interpretation of relationships between data points. Unlike k-means clustering, we do not need to specify the number of clusters. It is very efficient and can identify small clusters.
Implementation of Agglomerative Clustering in Python
We will use the iris dataset for demonstration. The first step is to import the necessary libraries and load the dataset.
import numpy as np import pandas as pd import matplotlib.pyplot as plt from sklearn.datasets import load_iris from sklearn.cluster import AgglomerativeClustering from scipy.cluster.hierarchy import dendrogram, linkage iris = load_iris() X = iris.data y = iris.target
The next step is to create a linkage matrix that contains the distances between each pair of clusters. We can use the linkage function from the scipy.cluster.hierarchy module to create the linkage matrix.
Z = linkage(X,'ward')
The ‘ward’ method is used to calculate the distances between the clusters. It minimizes the variance of the distances between the clusters being merged.
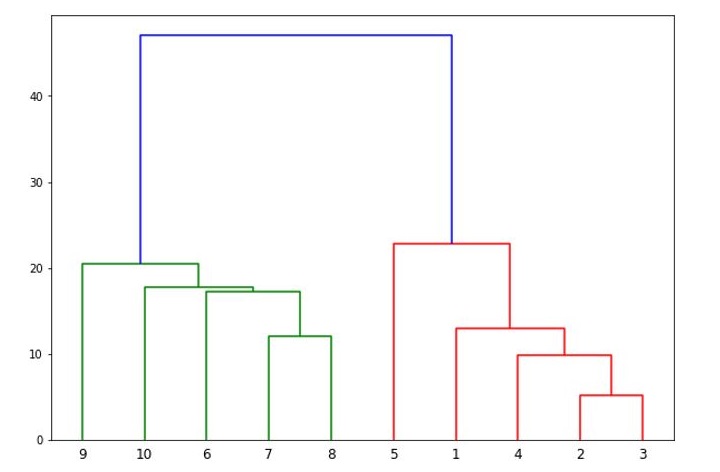
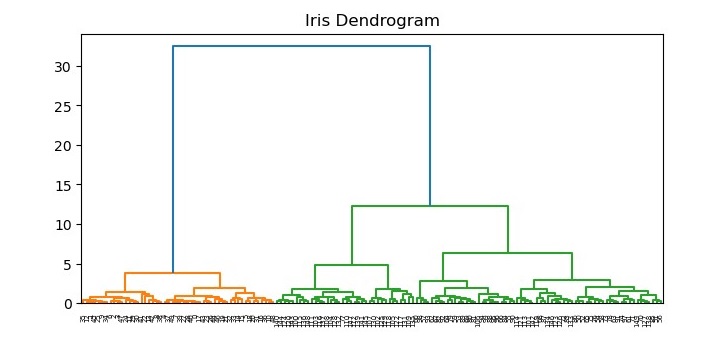
We can visualize the dendrogram using the dendrogram function from the same module.
plt.figure(figsize=(7.5,3.5))
plt.title("Iris Dendrogram")
dendrogram(Z)
plt.show()
The resulting dendrogram (see the following plot) shows the hierarchical relationship between the clusters. We can see that the algorithm has merged the closest clusters first, and the distance between the clusters increases as we move up the tree.
The final step is to apply the clustering algorithm and extract the cluster labels. We can use the AgglomerativeClustering class from the sklearn.cluster module to apply the algorithm.
model = AgglomerativeClustering(n_clusters=3) model.fit(X) labels = model.labels_
The n_clusters parameter specifies the number of clusters to be extracted from the data. In this case, we have specified n_clusters=3 because we know that the iris dataset has three classes.
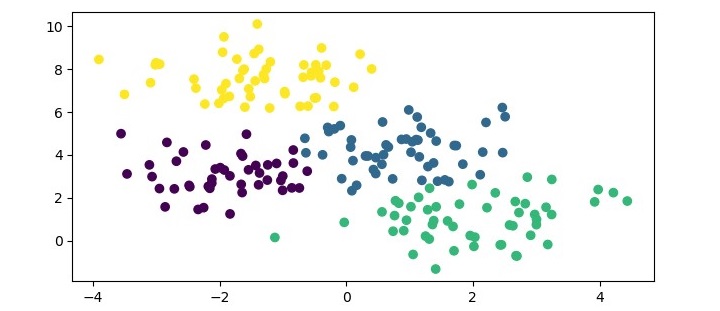
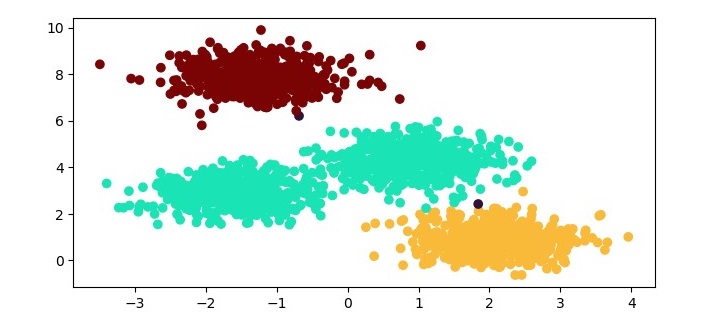
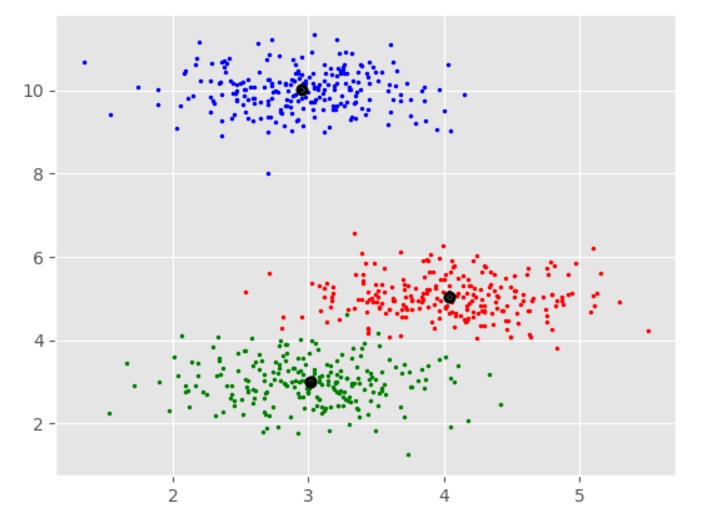
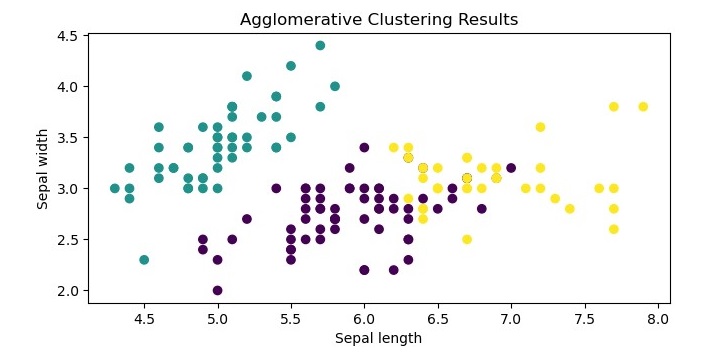
We can visualize the resulting clusters using a scatter plot.
plt.figure(figsize=(7.5,3.5))
plt.scatter(X[:,0], X[:,1], c=labels)
plt.xlabel("Sepal length")
plt.ylabel("Sepal width")
plt.title("Agglomerative Clustering Results")
plt.show()
The resulting plot shows the three clusters identified by the algorithm. We can see that the algorithm has successfully separated the data points into their respective classes.
Example
Here is the complete implementation of Agglomerative Clustering in Python −
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_iris
from sklearn.cluster import AgglomerativeClustering
from scipy.cluster.hierarchy import dendrogram, linkage
# Load the Iris dataset
iris = load_iris()
X = iris.data
y = iris.target
Z = linkage(X,'ward')# Plot the dendogram
plt.figure(figsize=(7.5,3.5))
plt.title("Iris Dendrogram")
dendrogram(Z)
plt.show()# create an instance of the AgglomerativeClustering class
model = AgglomerativeClustering(n_clusters=3)# fit the model to the dataset
model.fit(X)
labels = model.labels_
# Plot the results
plt.figure(figsize=(7.5,3.5))
plt.scatter(X[:,0], X[:,1], c=labels)
plt.xlabel("Sepal length")
plt.ylabel("Sepal width")
plt.title("Agglomerative Clustering Results")
plt.show()
Advantages of Agglomerative Clustering
Following are the advantages of using Agglomerative Clustering −
- Produces a dendrogram that shows the hierarchical relationship between the clusters.
- Can handle different types of distance metrics and linkage methods.
- Allows for a flexible number of clusters to be extracted from the data.
- Can handle large datasets with efficient implementations.
Disadvantages of Agglomerative Clustering
Following are some of the disadvantages of using Agglomerative Clustering −
- Can be computationally expensive for large datasets.
- Can produce imbalanced clusters if the distance metric or linkage method is not appropriate for the data.
- The final result may be sensitive to the choice of distance metric and linkage method used.
- The dendrogram may be difficult to interpret for large datasets with many clusters.
Applications of Agglomerative Clustering
You can find application of Agglomerative Clustering in many areas of unsupervised machine learning tasks. The following are some important areas of its applications in machine learning −
- Image Segmentation
- Document Clustering
- Customer Behaviour Analysis (Customer Segmentation)
- Market Segmentation
- Social Network Analysis